Understanding the processes that shape the community structure of Arctic phytoplankton is crucial for predicting responses of Arctic ecosystems to the ongoing ocean warming. In particular, little is known about the importance of phytoplankton dispersal by the North Atlantic Current and the prevalence and maintenance of Arctic endemism. We investigated the diversity and biogeography of diatoms from five Svalbard fjords and the Hausgarten observatory (Fram Strait) by combining diatom cultivation and 18S rRNA gene metabarcoding. In total, 50 diatom strains were isolated from the area during the HE492 cruise in August 2017. The strains were identified taxonomically using molecular and morphological approaches, and their biogeographic distribution was mapped using the local metabarcoding dataset and a global compilation of published metabarcoding datasets. Biogeographic analysis was also conducted for the locally most abundant diatom metabarcoding amplicon sequence variants. The biogeographic analyses demonstrated that Arctic diatoms exhibit three general biogeographic distribution types: Arctic, Arctic-temperate, and cosmopolitan. At Hausgarten and in outer Isfjorden on the west coast of Svalbard, the communities were dominated by genotypes with Arctic-temperate and cosmopolitan distribution. Diatom communities in nearby Van Mijenfjorden, inner Isfjorden and Kongsfjorden were dominated by genotypes with Arctic-temperate distribution, and cosmopolitan species were less abundant. The genotypes endemic to the Arctic had lower abundance on the west coast of Svalbard. The two northernmost fjords (Woodfjorden and Wijdefjorden) had a higher abundance of genotypes endemic to the Arctic. Our results demonstrate that the diatom communities in the Svalbard area consist of genotypes endemic to the Arctic, and genotypes with broader biogeographic distribution, all of which are further structured by local environmental gradients. Finer biogeographic patterns observed within Arctic-temperate and cosmopolitan genotypes suggest that certain genotypes can be used as indicators of increasing influence of Atlantic waters on the phytoplankton community structure in the Svalbard area.
Introduction
Diatoms are important primary producers in Arctic marine ecosystems. They dominate both pelagic and sympagic (ice-associated) primary production (Eamer et al., 2013) and have a crucial role in Arctic food web dynamics (Lovejoy et al., 2002) as well as the biogeochemical cycling of carbon (Le Moigne et al., 2015) and silica (Krause et al., 2018). With more than 1,000 described morphospecies (Poulin et al., 2011), Arctic diatoms exhibit a remarkable taxonomic diversity, along with a high degree of morphological, physiological and functional diversity (Fragoso et al., 2018). Accordingly, the abundance, community composition and distribution of diatoms in the Arctic realm ultimately shape ecosystem functioning (Wiedmann et al., 2020).
The Arctic today is a hotspot of climate change (Overland et al., 2019), with ocean warming, decreasing sea-ice cover (Kwok, 2018), and ocean freshening (Haine et al., 2015) likely to continue under projected carbon emission scenarios (Adakudlu et al., 2019). In this context, understanding the processes that are shaping Arctic diatom communities is essential for predicting the response of Arctic ecosystems to global warming. The structuring of Arctic diatom communities by seasonal and latitudinal patterns in light, nutrient availability, sea-ice and oceanographic conditions is generally well-understood (Eamer et al., 2013; Weydmann et al., 2013; Leu et al., 2015; Leeuwe et al., 2018). However, the fine-scale, genotype-level biogeographic patterns underlying the distribution of Arctic diatoms, particularly the relative importance of endemic Arctic species and of species that are dispersed with Atlantic circulation, are still unknown (Polyakov et al., 2017; Oziel et al., 2020).
Endemism is seemingly rare among marine protists, especially among planktonic species with large population sizes and the potential for global dispersal (Finlay, 2002). This paradigm has been challenged by evidence for endemism among marine planktonic protists, especially in polar areas (Darling et al., 2007; Lovejoy et al., 2007; Vyverman et al., 2010; Ribeiro et al., 2020; Yau et al., 2020). A certain degree of biogeographically restricted distribution has been reported for diatoms in the Arctic, notably for species associated with sea ice (Quillfeldt, 2000; Poulin et al., 2011). A recent, morphology-based analysis of the biogeography of selected cultured Arctic diatom species (Balzano et al., 2017) suggested that Arctic diatoms exhibit diverse biogeographic distribution patterns, which often extend beyond the Arctic realm into temperate and even warm latitudes.
In recent years, several studies have reported an apparent northward expansion of temperate phytoplankton, most notably bloom-forming haptophytes such as Emiliania huxleyi and Phaeocystis spp. (Orkney et al., 2020; Oziel et al., 2020), into the Atlantic sector of the Arctic. The phenomenon, termed “Atlantification”, has been correlated with the increasing inflow of warm Atlantic waters and warming of Arctic waters and is seen as an indicator of global warming (Polyakov et al., 2017; Neukermans et al., 2018). The relevance of dispersed temperate or low-latitude diatom species for community structuring in the Arctic is unknown. Based on available morphology-based data, cosmopolitan species constitute a significant portion of the Arctic diatom flora (Quillfeldt, 2000; Poulin et al., 2011). Unclear is whether they represent warm-water species dispersed with Atlantic circulation, truly cosmopolitan species also adapted to the Arctic environment, or cold-adapted cryptic species and genotypes restricted to the Arctic. Identifying these fine-scale biogeographic patterns is crucial for understanding processes that are shaping Arctic phytoplankton communities, and for developing tools for tracking changes in Arctic microalgal biodiversity related to the warming climate.
Exploring the biogeographic distribution of Arctic diatoms is challenging due to several methodological constraints. The morphology-based datasets are often limited by insufficient and inconsistent taxonomic identification. These issues are especially relevant for species complexes with cosmopolitan biogeographic distribution, which often include cryptic, cold-adapted species (Chamnansinp et al., 2013; Percopo et al., 2016) and cold-adapted genotypes (Stock et al., 2019) that are usually indistinguishable and easily overlooked in morphology-based surveys. On the other hand, combining morphological and molecular approaches (Balzano et al., 2017; Ribeiro et al., 2020) provides a more robust taxonomic framework for identifying Arctic diatoms and accounting for their cryptic diversity. When combined with ever-increasing environmental metabarcoding datasets, this approach allows for an in-depth analysis of the global distribution of morphologically-verified diatom genotypes using species-level molecular markers such as 18S rRNA gene sequences (De Luca et al., 2019).
This study aimed to reveal the diversity and biogeographic distribution of Arctic planktonic diatoms and understand the processes that shape the diatom community structure in the Svalbard area, particularly the interplay of dispersal by Atlantification, Arctic endemism and local environmental forcing at eastern Fram Strait and along five Svalbard fjords. The work followed three main lines: 1) morphological and molecular characterisation of cultured diatoms strains isolated from the Svalbard area; 2) exploration of the global and local distribution patterns of the cultured diatom genotypes and the most abundant metabarcoding ASVs using a global compilation of 18S V4 rRNA gene metabarcoding datasets; and 3) assessment of the contribution of genotypes with distinct biogeographic distribution to late-summer diatom communities on the west coast of Svalbard.
Material and methods
Studied area
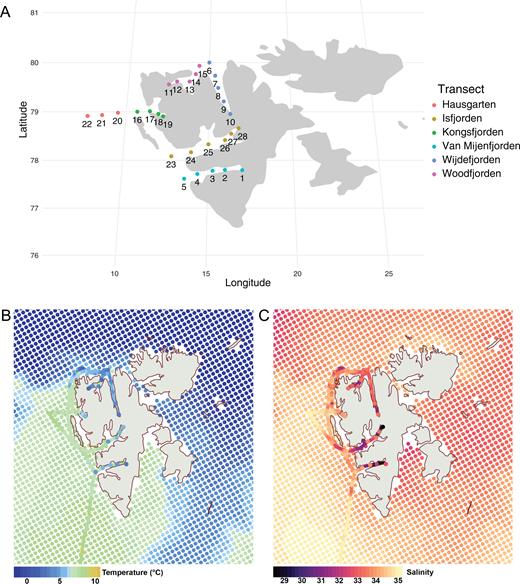
The samples were collected onboard R/V Heincke between July 29 and August 18, 2017, in the frame of the cruise number HE492 led by Alfred Wegener Institute (Germany). In total, 28 stations were sampled, forming transects of 4 to 6 stations within five Svalbard fjords (south to north: Van Mijenfjorden, Isfjorden, Kongsfjorden, Woodfjorden and Wijdefjorden) and a 3-station transect at the long-term ecological research observatory Hausgarten, located in eastern Fram Strait (Figure 1A).
Sampling stations and hydrographic conditions at the time of sampling. (A) Sampling stations and transects along the five Svalbard fjords and at Hausgarten area; (B) sea surface temperature and (C) salinity based on ferrybox and CTD data along with the satellite data derived from Copernicus ARCTIC_MULTIYEAR_PHY_002_003 (last access: 2022-06-17), averaged over 20170803-20170816 with the Europe coastline derived from the European Environment Agency (https://www.eea.europa.eu/ds_resolveuid/06227e40310045408ac8be0d469e1189) and the map shown in the NSIDC Sea Ice Polar Stereographic North projection using qGIS v3.24.2.
Sampling stations and hydrographic conditions at the time of sampling. (A) Sampling stations and transects along the five Svalbard fjords and at Hausgarten area; (B) sea surface temperature and (C) salinity based on ferrybox and CTD data along with the satellite data derived from Copernicus ARCTIC_MULTIYEAR_PHY_002_003 (last access: 2022-06-17), averaged over 20170803-20170816 with the Europe coastline derived from the European Environment Agency (https://www.eea.europa.eu/ds_resolveuid/06227e40310045408ac8be0d469e1189) and the map shown in the NSIDC Sea Ice Polar Stereographic North projection using qGIS v3.24.2.
The studied area is a unique natural laboratory for exploring how biogeographic processes and environmental gradients are shaping Arctic diatom communities (Figure 1B and C). The Hausgarten stations are located within the West Spitzbergen Current, an extension of the North Atlantic current that brings warmer and nutrient-rich Atlantic waters into the Arctic (Soltwedel et al., 2016). Isfjorden and Kongsfjorden, on the west coast of Svalbard, are increasingly influenced by inflowing Atlantic waters (Nilsen et al., 2016; Menze et al., 2020), as evidenced by the long-term trends in hydrography (Pavlov et al., 2013; Tverberg et al., 2019) and primary production (Hegseth and Tverberg, 2013). In contrast, the characteristic topography of the southernmost Van Mijenfjorden, with an island blocking the entrance to the fjord, reduces the inflow of coastal waters, and the fjord is consequently less influenced by the Atlantic water inflow (Skarðhamar and Svendsen, 2010; Nilsen et al., 2016). Finally, the two northernmost fjords, Wijdefjorden and Woodfjorden, are located at the northern edge of the West Spitzbergen Current and are the least affected by the North Atlantic Current of all the fjords studied here (Menze et al., 2020). Along with different levels of exposure to advection of warmer water masses, the studied fjords exhibit distinct environmental gradients defined by their geography and freshwater and nutrient input by glaciers and runoff (Cottier et al., 2010).
CTD profiling and water sampling
A conductivity-temperature-depth (CTD) probe (Seabird sbe911+, Seabird, USA) was deployed at each station to determine the hydrographical profile. The CTD was equipped with additional sensors measuring turbidity, fluorescence and oxygen levels. All CTD data collected during the cruise have been deposited in the Pangea data repository (https://doi.pangaea.de/10.1594/PANGAEA.881296). The CTD probe was equipped with a rosette with 10-L Niskin bottles, which were deployed to collect water samples from discrete depths after the initial onboard examination of the profile.
Culture isolation
Clonal cultures of planktonic diatoms were isolated from stations 6 and 7 located in Wijdefjorden and stations 20, 21 and 22 located at Hausgarten. A concentrated phytoplankton sample was collected from 0–30 m by a vertical net haul using a plankton net with mesh size of 20 µm. Upon arrival to deck the net haul sample was diluted to 1 L with filtered seawater, and 40 mL of this volume was transferred to a Nunclon culturing flask with added 10 mL of algal growth medium (IMR ½ medium with salinity 34; Eppley et al., 1967). In addition, water samples for metabarcoding collected by the Niskin bottles on the CTD from 3, 15, and 30 m were pooled. Some of this material was pre-filtered through a 200-µm mesh sieve and cells were collected on a 20-µm mesh sieve, rinsed off the sieve with seawater and transferred to a culturing flask to which was added 1/10 parts of IMR ½ medium with salinity 34. Both types of samples were stored onboard at 4ºC in an illuminated incubator until the isolation of cultures onboard and later in the lab at UiO.
The larger diatom species were obtained using capillary isolation. Concentrated samples were placed in a Petri dish and observed under a Zeiss Axiovert A1 inverted microscope (Zeiss, Oberkochen, Germany). Individual cells were isolated using a custom-made glass capillary connected to a rubber tube with a mouthpiece. Each cell was washed by repeated transfers to drops of filter-sterilized seawater and was finally transferred to an individual well on a 96-well culture plate (Thermo Fisher Scientific, USA) filled with approximately 300 µL of IMR ½ medium with salinity 34. Cells were kept in a culture room at 4ºC, under medium light intensity (approximately 10–20 μmol m–2 s–1) and 16 h/8 h light/dark cycle, and were inspected regularly for growth and contamination. When the presence of a single, exponentially growing culture was confirmed, the culture was transferred to a larger, 12-well culture plate (Thermo Fisher Scientific, USA) in 4 mL of the growth medium. For the smaller species (< 20 µm) dilution cultures were prepared from total integrated water samples collected by Niskin bottles (from depths of 3, 15 and 30 m), with a 6-step dilution series along the rows of the 96-well culture plate (Thermo Fisher Scientific, USA) with a final dilution of x106. Well plates were incubated in the same culture room as the capillary isolates, and the monocultures detected in the dilution series followed the same treatment as the capillary isolates.
When a culture reached high concentration in 4-mL wells, a drop (approximately 200 µL) of culture was inoculated into a glass cultivation tube with 10 mL medium and assigned a strain code. Subsamples of cultures were collected for live observation by light microscopy and imaging using a Zeiss Axioscope light microscope (Zeiss, Oberkochen, Germany) equipped with a Leica MC170 HD Camera (Leica, Germany). In addition, 1 mL of dense culture material was fixed with EM-grade glutaraldehyde (final concentration 1%; Electron Microscopy Sciences, Hatfield, PA, USA) and stored at 4ºC for future morphological analysis. Cultures were maintained in 10-mL glass tubes and transferred to a fresh medium every 5–7 weeks. Subsequently, cultures were deposited to the Norwegian Culture Collection of Algae (NORCCA), where they were assigned a new strain code and made publicly available (Table 1; www.norcca.scrol.net).
List of diatom strains isolated during this study
| . | GenBank Accession Number c . | |||
|---|---|---|---|---|
| Taxon . | Strain Code (NORCCA)a . | Stationb . | 18S . | 28S . |
| Bacillariaceae | ||||
| Cylindrotheca closterium | HE492-63 (UiO 469) | 7 (10) | OK147662 | OK147703 |
| Fragilariopsis sp.d | HE492-50 | 21 (28) | OK147664 | OK147701 |
| Pseudo-nitzschia granii | HE492-40 (UiO 453) | 20 (27) | OK147667 | OK147699 |
| Pseudo-nitzschia turgidula | HE492-37 (UiO 451) | 20 (27) | OK147665 | OK147698 |
| Pseudo-nitzschia turgidula | HE492-47 (UiO 460) | 21 (28) | OK147666 | OK147697 |
| Attheyaceae | ||||
| Attheya longicornis | HE492-59 (UiO 467) | 20 (27) | OK147651 | OK147726 |
| Cymatosiraceae | ||||
| Arcocellulus cornucervis | HE492-38 | 20 (27) | OK147650 | OK147729 |
| Arcocellulus cornucervis | HE492-43 (UiO 456) | 21 (28) | OK147649 | OK147728 |
| Arcocellulus cornucervis | HE492-71 (UiO 471) | 6 (9) | nae | OK147727 |
| Undetermined Cymatosiraceaed | HE492-46 (UiO 459) | 21 (28) | OK147663 | OK147702 |
| Chaetocerotaceae | ||||
| Chaetoceros contortus | HE492-06 (UiO 434) | 20 (27) | OK147652 | OK147725 |
| Chaetoceros convolutus | HE492-64 | 7 (10) | OK147653 | na |
| Chaetoceros debilis (Clade 1) | HE492-61 | 20 (27) | na | OK147722 |
| Chaetoceros debilis (Clade 2) | HE492-19 | 20 (27) | OK147654 | OK147724 |
| Chaetoceros debilis (Clade 2) | HE492-28 | 22 (29) | na | OK147723 |
| Chaetoceros diadema (Clade 1) | HE492-13 (UiO 440) | 20 (27) | na | OK147720 |
| Chaetoceros diadema (Clade 2)d | HE492-12 (UiO 439) | 20 (27) | OK147655 | OK147721 |
| Chaetoceros diadema (Clade 2)d | HE492-15 (UiO 441) | 20 (27) | na | OK147719 |
| Chaetoceros diadema (Clade 2)d | HE492-16 (UiO 442) | 20 (27) | na | OK147718 |
| Chaetoceros diadema (Clade 2)d | HE492-17 (UiO 443) | 20 (27) | na | OK147717 |
| Chaetoceros diadema (Clade 2)d | HE492-18 (UiO 444) | 20 (27) | OK147656 | OK147716 |
| Chaetoceros diadema (Clade 2)d | HE492-21 | 7 (10) | OK147657 | OK147715 |
| Chaetoceros diadema (Clade 2)d | HE492-58 (UiO 446) | 20 (27) | OK147658 | OK147714 |
| Chaetoceros diadema (Clade 2)d | HE492-65 | 22 (29) | na | OK147713 |
| Chaetoceros diadema (Clade 2)d | HE492-75 (UiO 475) | 20 (27) | na | OK147712 |
| Chaetoceros neogracilis (Clade 1) | HE492-73 (UiO 473) | 6 (9) | OK147659 | OK147711 |
| Chaetoceros sp.d | HE492-36 (UiO 450) | 6 (9) | OK147661 | OK147705 |
| Chaetoceros sp.d | HE492-02 | 20 (27) | na | OK147710 |
| Chaetoceros sp.d | HE492-03 | 20 (27) | OK147660 | OK147709 |
| Chaetoceros sp.d | HE492-05 (UiO 433) | 20 (27) | na | OK147708 |
| Chaetoceros sp.d | HE492-11 | 20 (27) | na | OK147707 |
| Chaetoceros sp.d | HE492-27 | 22 (29) | na | OK147706 |
| Leptocylindraceae | ||||
| Leptocylindrus sp. | HE492-14 | 20 (27) | na | OK147700 |
| Skeletonemataceae | ||||
| Skeletonema marinoi | HE492-39 (UiO 452) | 20 (27) | na | OK147691 |
| Skeletonema marinoi | HE492-42 (UiO 455) | 20 (27) | na | OK147690 |
| Thalassiosiraceae | ||||
| Shionodiscus bioculatus | HE492-10 (UiO 438) | 20 (27) | OK147668 | OK147696 |
| Shionodiscus bioculatus | HE492-23 | 7 (10) | OK147669 | OK147695 |
| Shionodiscus bioculatus | HE492-26 | 7 (10) | OK147670 | OK147694 |
| Shionodiscus bioculatus | HE492-31 | 6 (9) | OK147671 | OK147693 |
| Shionodiscus bioculatus | HE492-33 | 6 (9) | na | OK147692 |
| Thalassiosira gravida | HE492-07 (UiO 435) | 20 (27) | OK147672 | OK147689 |
| Thalassiosira gravida | HE492-08 (UiO 436) | 20 (27) | OK147673 | OK147688 |
| Thalassiosira gravida | HE492-30 (UiO 447) | 6 (9) | OK147674 | OK147687 |
| Thalassiosira gravida | HE492-32 (UiO 448) | 6 (9) | OK147675 | OK147686 |
| Thalassiosira gravida | HE492-34 | 6 (9) | OK147676 | OK147685 |
| Thalassiosira oceanica | HE492-54 | 22 (29) | OK147677 | OK147684 |
| Thalassiosira sp.d | HE492-45 (UiO 458) | 21 (28) | OK147678 | OK147683 |
| Thalassiosira sp.d | HE492-48 (UiO 461) | 21 (28) | OK147679 | OK147682 |
| Thalassiosira sp.d | HE492-52 (UiO 463) | 22 (29) | OK147680 | OK147681 |
| Corethraceae | ||||
| Corethron sp. | HE492-01 | 20 (27) | na | OK147704 |
| . | GenBank Accession Number c . | |||
|---|---|---|---|---|
| Taxon . | Strain Code (NORCCA)a . | Stationb . | 18S . | 28S . |
| Bacillariaceae | ||||
| Cylindrotheca closterium | HE492-63 (UiO 469) | 7 (10) | OK147662 | OK147703 |
| Fragilariopsis sp.d | HE492-50 | 21 (28) | OK147664 | OK147701 |
| Pseudo-nitzschia granii | HE492-40 (UiO 453) | 20 (27) | OK147667 | OK147699 |
| Pseudo-nitzschia turgidula | HE492-37 (UiO 451) | 20 (27) | OK147665 | OK147698 |
| Pseudo-nitzschia turgidula | HE492-47 (UiO 460) | 21 (28) | OK147666 | OK147697 |
| Attheyaceae | ||||
| Attheya longicornis | HE492-59 (UiO 467) | 20 (27) | OK147651 | OK147726 |
| Cymatosiraceae | ||||
| Arcocellulus cornucervis | HE492-38 | 20 (27) | OK147650 | OK147729 |
| Arcocellulus cornucervis | HE492-43 (UiO 456) | 21 (28) | OK147649 | OK147728 |
| Arcocellulus cornucervis | HE492-71 (UiO 471) | 6 (9) | nae | OK147727 |
| Undetermined Cymatosiraceaed | HE492-46 (UiO 459) | 21 (28) | OK147663 | OK147702 |
| Chaetocerotaceae | ||||
| Chaetoceros contortus | HE492-06 (UiO 434) | 20 (27) | OK147652 | OK147725 |
| Chaetoceros convolutus | HE492-64 | 7 (10) | OK147653 | na |
| Chaetoceros debilis (Clade 1) | HE492-61 | 20 (27) | na | OK147722 |
| Chaetoceros debilis (Clade 2) | HE492-19 | 20 (27) | OK147654 | OK147724 |
| Chaetoceros debilis (Clade 2) | HE492-28 | 22 (29) | na | OK147723 |
| Chaetoceros diadema (Clade 1) | HE492-13 (UiO 440) | 20 (27) | na | OK147720 |
| Chaetoceros diadema (Clade 2)d | HE492-12 (UiO 439) | 20 (27) | OK147655 | OK147721 |
| Chaetoceros diadema (Clade 2)d | HE492-15 (UiO 441) | 20 (27) | na | OK147719 |
| Chaetoceros diadema (Clade 2)d | HE492-16 (UiO 442) | 20 (27) | na | OK147718 |
| Chaetoceros diadema (Clade 2)d | HE492-17 (UiO 443) | 20 (27) | na | OK147717 |
| Chaetoceros diadema (Clade 2)d | HE492-18 (UiO 444) | 20 (27) | OK147656 | OK147716 |
| Chaetoceros diadema (Clade 2)d | HE492-21 | 7 (10) | OK147657 | OK147715 |
| Chaetoceros diadema (Clade 2)d | HE492-58 (UiO 446) | 20 (27) | OK147658 | OK147714 |
| Chaetoceros diadema (Clade 2)d | HE492-65 | 22 (29) | na | OK147713 |
| Chaetoceros diadema (Clade 2)d | HE492-75 (UiO 475) | 20 (27) | na | OK147712 |
| Chaetoceros neogracilis (Clade 1) | HE492-73 (UiO 473) | 6 (9) | OK147659 | OK147711 |
| Chaetoceros sp.d | HE492-36 (UiO 450) | 6 (9) | OK147661 | OK147705 |
| Chaetoceros sp.d | HE492-02 | 20 (27) | na | OK147710 |
| Chaetoceros sp.d | HE492-03 | 20 (27) | OK147660 | OK147709 |
| Chaetoceros sp.d | HE492-05 (UiO 433) | 20 (27) | na | OK147708 |
| Chaetoceros sp.d | HE492-11 | 20 (27) | na | OK147707 |
| Chaetoceros sp.d | HE492-27 | 22 (29) | na | OK147706 |
| Leptocylindraceae | ||||
| Leptocylindrus sp. | HE492-14 | 20 (27) | na | OK147700 |
| Skeletonemataceae | ||||
| Skeletonema marinoi | HE492-39 (UiO 452) | 20 (27) | na | OK147691 |
| Skeletonema marinoi | HE492-42 (UiO 455) | 20 (27) | na | OK147690 |
| Thalassiosiraceae | ||||
| Shionodiscus bioculatus | HE492-10 (UiO 438) | 20 (27) | OK147668 | OK147696 |
| Shionodiscus bioculatus | HE492-23 | 7 (10) | OK147669 | OK147695 |
| Shionodiscus bioculatus | HE492-26 | 7 (10) | OK147670 | OK147694 |
| Shionodiscus bioculatus | HE492-31 | 6 (9) | OK147671 | OK147693 |
| Shionodiscus bioculatus | HE492-33 | 6 (9) | na | OK147692 |
| Thalassiosira gravida | HE492-07 (UiO 435) | 20 (27) | OK147672 | OK147689 |
| Thalassiosira gravida | HE492-08 (UiO 436) | 20 (27) | OK147673 | OK147688 |
| Thalassiosira gravida | HE492-30 (UiO 447) | 6 (9) | OK147674 | OK147687 |
| Thalassiosira gravida | HE492-32 (UiO 448) | 6 (9) | OK147675 | OK147686 |
| Thalassiosira gravida | HE492-34 | 6 (9) | OK147676 | OK147685 |
| Thalassiosira oceanica | HE492-54 | 22 (29) | OK147677 | OK147684 |
| Thalassiosira sp.d | HE492-45 (UiO 458) | 21 (28) | OK147678 | OK147683 |
| Thalassiosira sp.d | HE492-48 (UiO 461) | 21 (28) | OK147679 | OK147682 |
| Thalassiosira sp.d | HE492-52 (UiO 463) | 22 (29) | OK147680 | OK147681 |
| Corethraceae | ||||
| Corethron sp. | HE492-01 | 20 (27) | na | OK147704 |
a Strains deposited in the Norwegian Culture Collection of Algae (NORCCA) have been assigned an additional code (starting with “UiO”).
b Station names in parentheses were used during the cruise and correspond to station names in the PANGEA publication (John and Wisotzki, 2017).
c All obtained 18S and 28S rRNA gene sequences have been deposited in the NCBI database and were assigned an accession number.
d Potentially novel undescribed species.
e Not available.
Molecular characterisation of cultures
The main aim of the sequencing effort was to obtain 18S and 28S rRNA gene sequences for all cultured genotypes, with the particular focus on sequencing the V4 region of the 18S rRNA gene, which was used for mapping the biogeographic distribution of cultured genotypes. A 1.5 mL volume of a dense culture was collected in an Eppendorf tube and centrifuged for 15 min at 6000 g (Eppendorf centrifuge 5424 R) to form a pellet. The supernatant was discarded, and the pellet was stored at –20ºC until DNA extraction, which was performed using Qiagen DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
For amplification of the 18S rRNA gene, the initial PCR reaction was run using 1F as a forward primer and 1528R (Medlin et al., 1988) as reverse primers (Table S1). The PCR reaction steps were as follows: initial denaturation (94ºC, 3.5 min), followed by 35 cycles of denaturation (94ºC, 50 s), annealing (55ºC, 50 s) and elongation (72ºC, 2 min), and the final elongation step (72ºC, 10 min). A variety of sequencing primers for the 18S rRNA gene was used (1F, 300R, 528F, 850F, 1055R, 1147R, and 1528R), depending on the strain (Table S1). For amplifying the partial 28S rRNA gene sequence, the D1R-F and D2C-R primer pair (Scholin et al., 1994; Table S1) was used in a PCR reaction with the following steps: initial denaturation at (94ºC, 2 min), then 35 cycles of denaturation (94ºC, 45 s), annealing (55ºC, 45 s) and elongation (72ºC, 1 min), followed by the final elongation step (72ºC, 10 min). The 28S rRNA gene fragment was sequenced using the same primers as in the PCR reaction. Sequencing data (sequences and chromatograms) were quality-checked, and contigs were assembled into consensus sequences using the De Novo Assemble tool, trimmed, edited and aligned in the Geneious software (v. 11.1.5). Sequences were submitted to NCBI, with accession numbers OK147649-147680 for 18S rRNA gene sequences, and OK147681-147729 for 28S rRNA gene sequences (Table 1).
Metabarcoding of field samples
A total of 60 L or 20 L of seawater were collected at three different depths with Niskin bottles (depths of 3 m, 15 m, and 30 m) and subsequently pooled. The 60 Lwere gravity-filtered over 200-µm and finally 20-µm mesh size sieves. For the smaller size fraction, after pre-filtration over 20 µm, an in-line filtration followed through 3-µm and 0.2-µm polycarbonate filters (Millipore, USA) with a Millipore Tripod unit (147-mm diameter; Millipore, USA) and a peristaltic pump during a maximum of 30 min. These filtrations yielded three size fractions: microplankton (20–200 µm), nanoplankton (3–20 µm), and picoplankton (0.2–3 µm). Filters were then added to warm lysis buffer, fast-frozen in liquid nitrogen, and kept at –80°C until further processing.
DNA extraction was performed with the NucleoSpin® Soil kit (Macherey-Nagel, Düren, Germany) in SL1 buffer according to the manufacturer’s protocol. The 18S rRNA gene V4 region was targeted for amplification using forward and reverse primers (Bradley et al., 2016; Table S2) with overhang adapters attached. The Illumina overhang nucleotide sequences were added to the 18S amplicon PCR forward and reverse primer. The workflow for preparation of 18S rRNA gene amplicons for the Illumina MiSeq system was derived from the “16S Metagenomic Sequencing Library Preparation B” document (part no.15044223; Rev.B) distributed by Illumina with modifications for preparation of 18S ribosomal gene amplicons. Paired end Illumina sequencing (MSC 2.5.0.5/RTA 1.18.54, 2 x 300 bp) of 152 plankton samples was performed on a MiSeq platform (Illumina, United States). The sequence data were processed with the DADA2 pipeline for detecting amplicon sequence variants (ASVs) in a sample from the library of noisy reads generated by amplicon sequencing (Rosen et al., 2012; Callahan et al., 2016) as described in Elferink et al. (2020).
For further analysis, all diatom ASVs (Class = “Bacillariophyta”; N = 230) were extracted from the original ASV table based on the taxonomy assigned during the bioinformatic processing by DADA2 pipeline using PR2 version 4.10.0 (https://github.com/pr2database/pr2database/releases/tag/4.10.0; Guillou et al., 2013) as the reference sequence database. The reduced, diatom-only ASV table and the corresponding taxonomical and environmental metadata were then incorporated into a phyloseq object (McMurdie and Holmes, 2013) in R used in all of the downstream analysis and visualisation of the metabarcoding data.
Phylogenetic analysis of diatom cultures
Sequences obtained from each of the 50 strains were first compared with the GenBank using the NCBI BLAST algorithm tool in Geneious (v. 11.1.5). After assigning the initial taxonomy based on the morphological observations and molecular (BLAST) results, the sequences of all the strains were aligned using the L-INS-i algorithm in mafft (v7.427; Stamatakis, 2014) to determine the number of unique 18S and 28S rRNA genotypes. For the 18S marker gene, the phylogenetic tree was prepared using selected morphologically verified reference sequences from PR2 (v. 4.10.0) and recently published Arctic-diatom reference sequences (Balzano et al., 2017; Ribeiro et al., 2020) and reference sequences of Chaetoceros from Gaonkar et al. (2018). In addition, 18S rRNA sequences of cultured genotypes were compared with the local metabarcoding dataset to identify ASVs that were 100% identical to these strains. These ASVs were added to the 18S rRNA alignment using -add fragments algorithm in mafft and included in the 18S rRNA gene phylogenetic tree. The alignment for the 18S rRNA gene tree included 32 sequences obtained from cultures in this study, 17 ASVs from the local metabarcoding dataset and 81 reference sequences (a total of 130 sequences and 2084 nucleotide positions). For the 28S rRNA tree, we used morphologically verified reference sequences from GenBank (a total of 117 sequences with 700 positions). For both marker genes, sequences were aligned using the L-INS-i algorithm in mafft, and Maximum Likelihood phylogenetic trees were made using RAxML (v8.2.12; GTRGAMMA model with 1000 bootstrap replications).
Phylogenetic placement of diatom ASVs
To determine the phylogenetic placement of cultured genotypes and diatom ASVs obtained through metabarcoding, we compiled a diatom reference library with an improved representation of Arctic taxa. The diatom sequences (class==“Bacillariophyceae”) were extracted from the PR2 database (v.4.10.0; Guillou et al., 2013) and all the environmental (“ENV”) diatom sequences were removed, leaving the total of 1965 sequences. To these sequences, we added 218 recently published 18S rRNA reference sequences of the genus Chaetoceros (Gaonkar et al., 2018), novel morphologically verified Arctic diatom reference sequences published by Balzano et al. (2017) (19 sequences) and Ribeiro et al. (2020) (33 sequences) (all recently included in the PR2 v. 4.14.0), as well as 32 sequences newly obtained in this study. The initial reference alignment of the 2267 reference diatom sequences and three outgroup sequences (belonging to class Bolidophyceae, genus Triparma) was created using the FFT-NS-i algorithm with a maximum of 1000 iterations in mafft. After inspection of the alignment and trimming of the ends extending beyond the 1F-1528R primer positions (Medlin et al., 1988), the 230 diatom ASVs obtained in this study were placed in the alignment using the -add fragments algorithm in mafft. The final alignment containing reference sequences, our cultured genotypes and ASVs was used to build a Maximum Likelihood phylogenetic tree using the RAxML program (GTRGAMMA model with 1000 bootstrap replications). The final phylogenetic tree (Figure S1; interactive tree provided in ITOL: https://itol.embl.de/tree/193157255105100381657613640) was then used to assign a phylogeny-based taxonomic identity at the lowest possible taxonomic level to each of the diatom ASVs, following best practice and guidelines for the phylogeny-based taxonomic assignment (Zimmermann et al., 2015; Gaonkar et al., 2020).
Biogeographic analysis
For biogeographic analysis, we used a compilation of published metabarcoding datasets from different surveys that employed environmental metabarcoding targeting the V4 region of the 18S rRNA gene. The compiled dataset (metaPR2; Vaulot et al., 2022) has global coverage of coastal areas as it includes the samples from the Ocean Sampling Day survey (Kopf et al., 2015), notably in the northern hemisphere, as well as several oceanic transects based on the metabarcoding data collected within, e.g. the Tara Ocean (de Vargas et al., 2015) and Malaspina expeditions (Duarte, 2015). The dataset has a good coverage of the Arctic realm, especially the Atlantic sector and Greenland, with both planktonic and sea-ice habitats. List of all datasets included in the compiled dataset and related metadata are provided in the GitHub repository (https://github.com/vaulot/metapr2_HE492). The map of the sampling stations included in the combined dataset is shown in Figures 2–4.
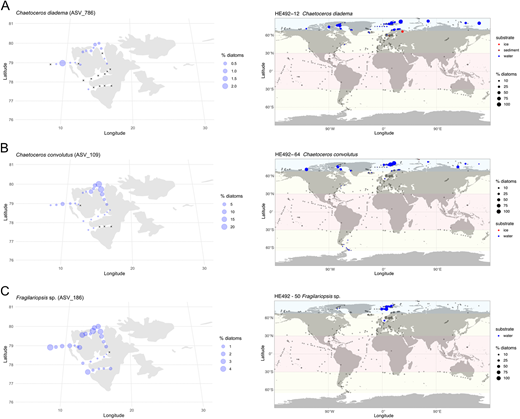
Examples of genotypes with Arctic biogeographic distribution. Maps showing examples of Arctic biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Chaetoceros diadema (ASV_786); (B) Chaetoceros convolutus (ASV_109); and (C) Fragilariopsis sp. (ASV_186). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
Examples of genotypes with Arctic biogeographic distribution. Maps showing examples of Arctic biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Chaetoceros diadema (ASV_786); (B) Chaetoceros convolutus (ASV_109); and (C) Fragilariopsis sp. (ASV_186). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
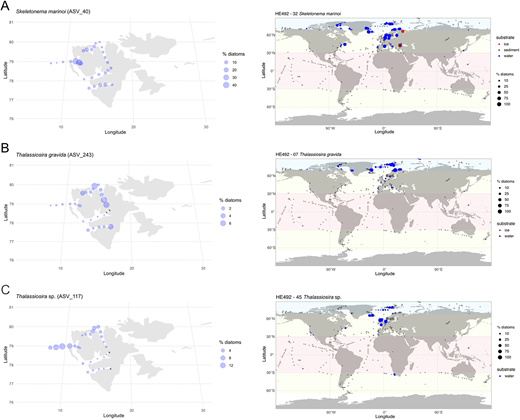
Examples of genotypes with Arctic-temperate biogeographic distribution. Maps showing examples of Arctic-temperate biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Skeletonema marinoi (ASV_40); (B) Thalassiosira gravida (ASV_243); and (C) Thalassiosira sp. (ASV_117). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
Examples of genotypes with Arctic-temperate biogeographic distribution. Maps showing examples of Arctic-temperate biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Skeletonema marinoi (ASV_40); (B) Thalassiosira gravida (ASV_243); and (C) Thalassiosira sp. (ASV_117). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
Examples of genotypes with cosmopolitan biogeographic distribution. Maps showing examples of cosmopolitan biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Shionodiscus bioculatus (ASV_110); (B) Pseudo-nitzschia granii (ASV_54); and (C) Thalassiosira oceanica (ASV_187). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
Examples of genotypes with cosmopolitan biogeographic distribution. Maps showing examples of cosmopolitan biogeographic distribution based on the metaPR2 database (right) along with distribution of example genotypes in the local metabarcoding dataset (left): (A) Shionodiscus bioculatus (ASV_110); (B) Pseudo-nitzschia granii (ASV_54); and (C) Thalassiosira oceanica (ASV_187). Symbol size is related to the relative abundance of genotype in respect to total diatom reads within each sample of the compiled metabarcoding dataset. Cross symbol (+) indicates samples in which the genotypes were not found.
Metadata and sequence processing are detailed in Vaulot et al. (2022). In brief, raw sequence files and metadata were downloaded from NCBI SRA web site (https://www.ncbi.nlm.nih.gov/Traces/studies) when available or obtained directly from the investigators. For each study, raw sequences files were processed independently de novo. Primer sequences were removed using cutadapt (Martin, 2011) and amplicon processing was performed under the R software (R 2013) using the DADA2 package (Callahan et al., 2016). Taxonomic assignation of ASVs was performed against the PR2 database (Guillou et al., 2013) version 4.12 (https://pr2-database.org; https://github.com/pr2database/pr2database/releases/tag/v4.12.0). ASV information was retrieved from final database available at https://shiny.metapr2.org.
For the biogeographic analysis of cultured genotypes, the 18S rRNA gene sequences of each genotype were compared with the ASVs found in the compiled dataset using the local BLAST tool in Geneious. After finding the ASVs in the dataset that were identical (100% pairwise identity) to the cultured genotypes, their distribution was mapped using an R script (available at https://vaulot.github.io/metapr2_HE492/metaPR2_HE492_diatoms.html), along with the relative contribution of each ASV to diatom assemblage at the sampling site. The same procedure was followed for mapping the distribution of the most abundant ASVs from the local metabarcoding dataset. After producing and examining the distribution maps, each genotype or ASV from the local metabarcoding dataset was assigned a descriptive biogeographic distribution type based on its global distribution pattern (Tables 2 and S3), namely Arctic (Figure 2), Arctic-temperate (Figure 3) or cosmopolitan (Figure 4).
Descriptions of biogeographic distribution types as defined in this study
| Biogeographic Distribution Type . | Description . |
|---|---|
| Arctic | Genotype has a biogeographic distribution mostly restricted to the Arctic circle (above 66ºN; Figure 2). |
| Arctic-temperate | Genotype is present in the Arctic and at temperate latitudes (30ºN–66ºN and 30ºS–66ºS; Figure 3). |
| Cosmopolitan | Genotype is present in the Arctic and has a cosmopolitan distribution (Figure 4). |
| Biogeographic Distribution Type . | Description . |
|---|---|
| Arctic | Genotype has a biogeographic distribution mostly restricted to the Arctic circle (above 66ºN; Figure 2). |
| Arctic-temperate | Genotype is present in the Arctic and at temperate latitudes (30ºN–66ºN and 30ºS–66ºS; Figure 3). |
| Cosmopolitan | Genotype is present in the Arctic and has a cosmopolitan distribution (Figure 4). |
Results
Overview of cultured diatom strains
In total, 50 diatom strains were isolated from Hausgarten (37 strains) and Wijdefjorden (13 strains) stations, most of them belonging to the families Chaetocerotaceae (22 strains) and Thalassiosiraceae (14 strains; Table 1). The remaining strains were isolated from families Bacillariaceae (5 strains), Cymatosiraceae (4 strains) and Skeletonemataceae (2 strains), while Attheyaceae, Leptocylindraceae and Corethraceae were each represented with a single strain. The molecular analysis yielded a total of 49 28S rRNA gene sequences and 32 18S rRNA gene sequences. The phylogenetic analysis confirmed that the strains belonged to 23 distinct diatom genotypes. At least one marker gene was sequenced for each genotype, and the V4 region of the 18S rRNA gene was sequenced for 19 distinct genotypes (Table 1).
Taxonomy and biogeographic distribution patterns of cultured diatom genotypes
Family Bacillariaceae
Cylindrotheca closterium strain HE492-63 (Wijdefjorden). Cells solitary, elongated, with two chloroplasts. Typical widening of the central part of the valve (Figure 5A). 18S rRNA gene sequence without a 100% match in GenBank (Figure 6). A 1 bp difference in the 18S rRNA gene sequence compared with the closest cultured strain KMMCC B-181 (South Korea). Sister clade to recently sequenced Arctic strains RCC5303, RCC5206 (Ribeiro et al., 2020) and RCC1985 (Balzano et al., 2017) (Figures 6 and S1). The 28S rRNA gene sequence identical to Strain KD10 (Kongsfjorden, Svalbard), a cold-adapted ecotype (Stock et al., 2019; Figure 7). Genotype found both in the local (ASV_766) and in the compiled dataset. Patchy local distribution. Higher read abundances in the innermost parts of all fjords except Isfjorden. Absent from the outermost Hausgarten stations (Figure S2A). Cosmopolitan distribution. Common in the Arctic realm, including sea-ice communities in the Arctic Basin (Figure S2A). The 28S rRNA signature suggests an Arctic, cold-adapted population or a cryptic Arctic genotype that cannot be distinguished using the 18S rRNA V4 marker and light microscopy (LM)-based morphology.
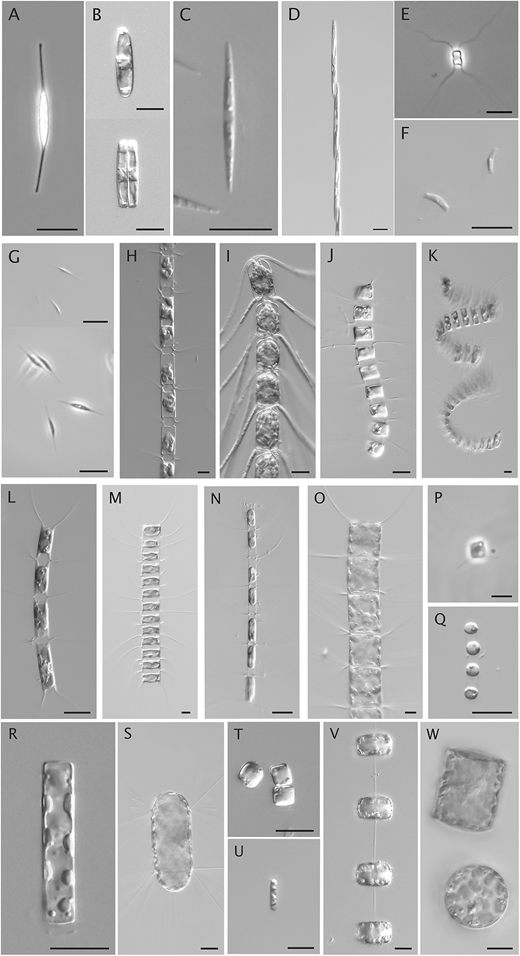
Light microscopy images of cultured genotypes. Scale bar = 10 µm in all images. (A) Cylindrotheca closterium (HE492-63); (B) Fragilariopsis sp. (HE492-50); (C) Pseudo-nitzschia granii (HE492-40); (D) Pseudo-nitzschia turgidula (HE492-37); (E) Attheya longicornis (HE492-59); (F) Arcocellulus cornucervis (HE492-43); (G) undetermined Cymatosiraceae (HE492-46); (H) Chaetoceros contortus (HE492-06); (I) Chaetoceros convolutus (HE492-64); (J) Chaetoceros debilis Clade 1 (HE492-61); (K) Chaetoceros debilis Clade 2 (HE492-28); (L) Chaetoceros diadema Clade 1 (HE492-13); (M) Chaetoceros diadema Clade 2 (HE492-17); (N) Chaetoceros sp. (HE492-05); (O) Chaetoceros sp. (HE492-36); (P) Chaetoceros neogracilis (HE492-73); (Q) Skeletonema marinoi (HE492-42); (R) Leptocylindrus sp. (HE492-14); (S) Corethron sp. (HE492-01); (T) Thalassiosira oceanica (HE492-54); (U) Thalassiosira sp. (HE492-45); (V) Thalassiosira gravida (HE492-07); and (W) Shionodiscus bioculatus (HE492-23).
Light microscopy images of cultured genotypes. Scale bar = 10 µm in all images. (A) Cylindrotheca closterium (HE492-63); (B) Fragilariopsis sp. (HE492-50); (C) Pseudo-nitzschia granii (HE492-40); (D) Pseudo-nitzschia turgidula (HE492-37); (E) Attheya longicornis (HE492-59); (F) Arcocellulus cornucervis (HE492-43); (G) undetermined Cymatosiraceae (HE492-46); (H) Chaetoceros contortus (HE492-06); (I) Chaetoceros convolutus (HE492-64); (J) Chaetoceros debilis Clade 1 (HE492-61); (K) Chaetoceros debilis Clade 2 (HE492-28); (L) Chaetoceros diadema Clade 1 (HE492-13); (M) Chaetoceros diadema Clade 2 (HE492-17); (N) Chaetoceros sp. (HE492-05); (O) Chaetoceros sp. (HE492-36); (P) Chaetoceros neogracilis (HE492-73); (Q) Skeletonema marinoi (HE492-42); (R) Leptocylindrus sp. (HE492-14); (S) Corethron sp. (HE492-01); (T) Thalassiosira oceanica (HE492-54); (U) Thalassiosira sp. (HE492-45); (V) Thalassiosira gravida (HE492-07); and (W) Shionodiscus bioculatus (HE492-23).
The 18S rRNA phylogeny of cultured strains. Maximum-likelihood tree was generated in RAxML using the GTRGAMMAI model with 1000 bootstrap replications. Only bootstrap values > 70 are shown. The cultured strains are marked in red, ASVs obtained from the local metabarcoding dataset are marked in blue, and reference diatom sequences are marked in black. Numbers next to ASVs indicate their identifier and total read abundance in the metabarcoding dataset.
The 18S rRNA phylogeny of cultured strains. Maximum-likelihood tree was generated in RAxML using the GTRGAMMAI model with 1000 bootstrap replications. Only bootstrap values > 70 are shown. The cultured strains are marked in red, ASVs obtained from the local metabarcoding dataset are marked in blue, and reference diatom sequences are marked in black. Numbers next to ASVs indicate their identifier and total read abundance in the metabarcoding dataset.
The 28S rRNA phylogeny of cultured strains. Maximum-likelihood tree was generated in RAxML using the GTRGAMMAI model with 1000 bootstrap replications. Only bootstrap values > 70 are shown. The cultured strains are marked in red, and reference diatom sequences are marked in black.
The 28S rRNA phylogeny of cultured strains. Maximum-likelihood tree was generated in RAxML using the GTRGAMMAI model with 1000 bootstrap replications. Only bootstrap values > 70 are shown. The cultured strains are marked in red, and reference diatom sequences are marked in black.
Fragilariopsis sp. strain HE492-50 (Hausgarten). Cells solitary, not chain-forming, with two chloroplasts (Figure 5B). Valve lanceolate in trans-valval view and rectangular in girdle view. Characteristic row of costae clearly visible at the valve margin. The LM morphology insufficient for species-level identification. No 100% match in GenBank for either of the two markers. Most similar to various Fragilariopsis spp. and Neodenticula seminae (based on 28S rRNA; Figure 7) sequences. The ASV_186 matches the V4 sequence of this genotype, forming a clade with two more fairly abundant ASVs likely representing other unidentified Fragilariopsis species (Figure S1). ASV ubiquitous within the local dataset. Abundant in the northern parts of the studied area (Figure S2B). Globally, restricted to Arctic. No records in the Arctic Basin or in sea-ice samples.
Pseudo-nitzschia granii strain HE492-40 (Hausgarten). Relatively small, lanceolate cells, long chains with overlapping valve-tips, often solitary in culture (Figure 5C). Both 18S rRNA (Figure 6) and 28S rRNA (Figure 7) identical to sequences described by Balzano et al. (2017) from Beaufort Sea and UNC 1102 from North West Pacific. A 100% match with ASV_54. Relatively abundant at all stations in the local dataset (Figure S2C). Globally cosmopolitan distribution in coastal and oceanic areas. Common in the Atlantic sector of the Arctic and in western Greenland, but not in the Arctic Basin.
Pseudo-nitzschia turgidula strains HE492-37 and HE492-47 (Hausgarten). Cells relatively large, lanceolate. Chains typical for the genus Pseudo-nitzschia (Figure 5D). First record of this species in the Arctic. The 28S rRNA gene sequence identical to strain NWFSC-255 (Northeastern Pacific Ocean; Lundholm et al., 2012; Figure 7) identified as P-n. cf. turgidula. Two 100% matching 18S rRNA gene sequences (unknown origin) in the PR2 reference database identified as P-n. turgidula (Figures 6 and S1). Matching ASV_179 most abundant at Hausgarten and present in Isfjorden and Kongsfjorden. Mostly absent from other fjords (Figure S2D). Globally, cosmopolitan but rare. Tropical coastal and oceanic stations and southern temperate and cold areas. Absent from the Arctic Basin. Could represent a warm-water genotype dispersed with Atlantic circulation.
Attheya longicornis strain HE492-59 (Hausgarten). Cells solitary, rectangular. Long undulating spines typical for this species projecting outwards diagonally from each valve end (Figure 5E). Phylogenetic placement within the Attheya clade. Unresolved distinction between A. septentrionalis and A. longicornis, possibly due to morphological misidentification of reference sequences. The 18S rRNA gene sequence identical to RCC2042 (Beaufort Sea; Balzano et al., 2017) and UNBF-P25C1 (Bay of Fundy, North Atlantic) identified as A. septentrionalis (Figures 6 and S1). The 28S rRNA gene sequence matches several GenBank entries, including the above-mentioned RCC2042 from the Arctic (Figure 7). Matching ASV_570 is the most abundant ASV from the Attheya clade in the local metabarcoding dataset. Abundant in Woodfjoden and Wijdefjorden, rare in the southern fjords and Hausgarten (Figure S2E). Arctic-temperate distribution. Clear Arctic presence including sea-ice samples.
Family Cymatosiraceae
Arcocellulus cornucervis strains HE492-38, -43 (Hausgarten) and HE492-71 (Wijdefjord). Saddle-like cells in girdle view. Characteristic pili extending inwards from the valve ends (Figure 5F). 18S (Figure 6) and 28S rRNA (Figure 7) gene sequences identical to strain RCC2270 (Beaufort Sea; Balzano et al., 2017), strain RCC5530 (Baffin Bay; Ribeiro et al., 2020), and strain NIES-3970 identified as Minutocellus polymorphus (unknown origin). In the reference phylogenetic tree, the 18S rRNA gene sequences and four related ASVs form a well-supported clade, indicating additional genotypes within this species present in the studied area (Figure S1). The most abundant ASV (ASV_24) from the Arcocellulus clade identical to the cultured strains. Widely distributed. Higher abundance in the southern fjords (Van Mijendfjorden, Isfjorden). Lower abundance in the northern fjords and at Hausgarten (Figure S3A). Arctic-temperate distribution. Present in Atlantic coastal areas, the Atlantic sector of the Arctic, and the Arctic Basin including sea-ice communities. Relatively rare beyond the polar front.
Unidentified strain belonging to the Cymatosiraceae family, HE492-46 (Hausgarten). Cells solitary. Elongated frustules with pointed ends, asymmetrical widening of valve along the apical axis (Figure 5G). Both 18S and 28S rRNA gene sequences have no match in GenBank (Figures 6 and 7). Novel clade within the Cymatosiraceae family. Three distinct ASVs from local metabarcoding dataset placed in the clade (Figure S1). ASV_46 identical to the cultured strain. Wide distribution in all fjords. Highest read abundance in Isfjorden (Figure S3B). Arctic-temperate distribution. Both coastal and open ocean presence. Not found in the Arctic Basin or the sea-ice samples.
Family Chaetocerotaceae
Chaetoceros contortus strain HE492-06 (Hausgarten). Cells rectangular. Straight chains with setae emerging closer to the centre of the valve. Fused interlocking setae of neighbouring cells forming wide, hexagonal apertures (Figure 5H). Some apertures in a chain rotated by 90 degrees, dividing chains into segments with differently rotated apertures. Both 18S and 28S rRNA gene phylogeny placing the strain into the C. contortus clade. Both 28S and 18S rRNA gene sequences identical to strains Ch9A2, Ch12A4, Ch8A1 and Ch2B2 (Chile waters; Figures 6 and 7). Six ASVs found in the studied area placed in the C. contortus clade. Most abundant ASV_1089 identical to the cultured genotype (Figure S1). Higher read abundance in the inner parts of the northern fjords. Lower abundance in the outer Hausgarten stations (Figure S3C). Arctic-temperate distribution, prevalent in coastal samples.
Chaetoceros convolutus strain HE492-64 (Wijdefjorden). Large, heavily silicified species. Cells rectangular. Convex anterior valve, longer in the pervalvar axis than the apical axis. Chloroplasts in setae, tightly interlocking neighbouring cells leaving almost no apertures (Figure 5I). Chains slightly curved and twisted, the posterior terminal valve often has one large seta. No 100% matching 18S rRNA gene sequences in GenBank (Figure 6). Phylogenetical placement with other heavily silicified Chaetoceros species with chloroplasts in setae (subgenus Phaeoceros) such as C. peruvianus and C. rostratus (Figure S1). Matching ASV_109 abundant in the northern fjords and at Hausgarten. Absent in the inner Isfjord and Van Mijenfjorden (Figure S3D). Predominantly found in the high Arctic, but also present in the Antarctic.
Chaetoceros debilis strain HE492-61(Hausgarten) belongs to Clade 1 and strains HE492-19 and -28 (Hausgarten) to Clade 2, based on 18S and 28S rRNA gene phylogenies (Figures 6 and 7), following Gaonkar et al. (2018). The Clade 1 strain has rectangular cells, almost identical length of pervalvar and apical axis. Thin setae emerging from the corners of the cells, slightly closer to the valve centre, fusing with neighbouring setae to form a wide hexagonal aperture (Figure 5J). Chains gently curved, spiralling and twisting along the chain axis. The Clade 2 strains have rectangular cells, shorter pervalvar axis compared with the apical axis, relatively narrow apertures formed by fusion of long basal parts of neighbouring setae (Figure 5K). In contrast to Clade 1 strain, the chains have pronounced spiral shape and are twisted along the chain axis. Despite isolating two genotypes based on the 28S rRNA gene marker, a single ASV from this clade was found in the metabarcoding dataset, with identical sequence to the Clade 1 genotype (Figure S1). Locally high abundance at all stations except Isfjorden (Figure S3E). Arctic-temperate, including Arctic Basin.
Two genotypes of Chaetoceros diadema were isolated. Strain HE492-13 (Hausgarten) is placed in Clade 1 of this species based on the 28S rRNA phylogeny. The other nine strains (HE492-12, -15, -16, -17, -18, -21, -58, -65, -and -75), all but one (HE492-21 from Wijdefjorden) isolated at Hausgarten, form a new clade within the C. diadema species complex, based on both 28S and 18S rRNA gene phylogeny (Gaonkar et al., 2018; Figures 6 and 7). The Clade 1 strain has rectangular cells, pervalvar axis approximately twice as long as the apical axis (Figure 5L). Relatively long basal parts of the setae emerging from the corners of the cells, interlocking neighbouring setae forming rectangular, wide apertures. Chains relatively short, gently curved. Terminal setae extending diagonally outwards from the short basal parts. Short convex protuberance on the terminal valves. The novel clade has rectangular cells, apical axis longer than the pervalvar axis, as opposed to Clade 1 (Figure 5M). Valves slightly convex. Basal parts of neighbouring setae interlocking to form narrow windows. Chains relatively short and straight. Terminal setae projecting outwards diagonally then running parallel to each other. A single ASV_786 matching the novel clade was retrieved in the local metabarcoding dataset. Predominantly in the northern parts of the studied area (Figure S4A). Globally, mostly restricted to the Arctic, few minor observations below the Arctic circle.
Chaetoceros neogracilis strain HE492-73 (Wijdefjorden). Small single cells, rectangular (Figure 5P), short straight setae emerging diagonally from the corners of valves. 18S (Figure 6) and 28S rRNA gene sequences (Figure 7) placed into Clade 1 of the C. neogracilis complex (Balzano et al., 2017). Three ASVs from the local metabarcoding dataset placed in the C. neogracilis clade. ASV_121 identical to cultured genotype, but also to the Clade 2 reference sequences (Figure S1), such that the V4 region is identical for the two clades. Present at all stations. Highest abundance in Van Mijenfjorden in the south and the two northern fjords. Lower abundance at Hausgarten, Kongsfjorden and Isfjorden (Figure S4B). Arctic-temperate distribution. Abundant in the Arctic, including Arctic Basin and sea-ice.
Chaetoceros sp. strain HE492-36 (Wijdefjorden). Morphologically similar to C. lauderi and C. teres. Cells rectangular. Pervalval and apical axes of equal lengths (Figure 5N). Cells densely connected in the chain, overlapping setae, almost no apertures. Terminal setae extending outwards with a short basal part. Chains straight, not twisted along the chain axis. The 28S rRNA gene sequence has no 100% match in GenBank (Figure 7). The 18S rRNA gene sequence 99.9% identical to the strain CCAP 1010/16 (unknown origin; Figure 6). The phylogenetic placement indicates an undescribed, novel species of Chaetoceros affiliated to the C. brevis/C. teres/C. lauderi clade, with the highest similarity to C. lauderi (Figure S1). A single ASV_1173 matched the cultured strain. Very rare, most abundant in the northern fjords and at two Hausgarten stations. Arctic-temperate distribution (Figure S4C).
Chaetoceros sp. strains HE492-02, -03, -05, -11 and -27 (Hausgarten). A novel species and a new clade within the Chaetoceros genus. Cells rectangular and elongated (Figure 5O). Setae from neighbouring cells merging at the relatively short basal parts, leaving small, elliptical apertures. Chains straight, long, terminal setae extending almost laterally in respect to the chain axis. The 28S rRNA marker gene has no 100% match in the GenBank. The 18S rRNA sequence identical to a Chaetoceros sp. strain (coast of Japan; Figures 6 and 7). Phylogenetic placement as a sister group to the C. anastomosans/C. vixvisibilis clade, though with low bootstrap support (Figure S1). ASV_1516 identical to cultured strains, Only detected at Hausgarten (Figure S4D). Arctic-temperate distribution. Rare in the Arctic Basin.
Family Skeletonemataceae
Skeletonema marinoi, strains HE492-39 and HE492-42 (Hausgarten). Cells relatively small, rectangular in girdle view, connected with interlocking processes growing from the edges of valves, forming long characteristic chains (Figure 5Q). Only 28S rRNA gene sequences obtained, identical to S. marinoi (Figure 7). A single ASV (ASV_40) belonging to Skeletonema marinoi was found in the metabarcoding dataset, but not certain is whether it matches the 18S rRNA gene sequences of the cultured genotype, as it may represent other Skeletonema species, too (Figure S1). Read abundance highest in Kongsfjorden, but high in other fjords as well. Slightly lower abundance at Hausgarten (Figure S4E). Globally very common, Arctic-temperate distribution, including sea-ice samples.
Family Thalassiosiraceae
Shionodiscus bioculatus strains HE492-10, -23, -26, -31 and -33 (Hausgarten and Wijdefjorden). Cells cylindrical, often solitary, sometimes forming shorter chains of cells closely linked with numerous organic threads (Figure 5W). The 18S (Figure 6) and 28S (Figure 7) rRNA marker genes all match S. bioculatus strains RCC5532 and RCC1991 (Beaufort Sea and Baffin Bay, respectively; Balzano et al., 2017). ASV_110 had an identical sequence to the cultured strain. Present at all stations. Higher abundance at Hausgarten and outer parts of fjords (Figure S5A). Cosmopolitan, including high Arctic and Antarctic.
Thalassiosira gravida strains HE492-07, -08, -30, -32 and -34 (Hausgarten and Wijdefjorden). Cells chain-forming, cylindrical, rectangular in girdle view with slightly curved edges of valves (Figure 5V). Single thick thread connecting cells in a chain. Numerous strutted processes visible in girdle view. Both 18S (Figures 6 and S1) and 28S (Figure 7) rRNA marker genes match T. gravida and T. rotula genotypes. These were considered the same species with temperature-induced morphological variability (Sar et al., 2011), though some studies suggest that they are genetically distinct based on their ITS sequences (Whittaker et al., 2012). A single ASV_243 was identical to the cultured strains. Present at all stations. Most abundant in the innermost parts of Woodfjorden, Wijdefjorden and Van Mijenfjorden (Figure S5B). Arctic-temperate distribution, few records in Arctic Basin.
Thalassiosira oceanica strain HE492-54 (Hausgarten). Small and cylindrical, mostly solitary cells, slightly rounded valve edges in girdle view (Figure 5T). The 18S (Figure 6) and 28S (Figure 7) rRNA marker genes match previously sequenced representatives of this species. Phylogenetic placement in 18S rRNA tree is within a well-supported T. oceanica clade (Figure S1). The ASV_187 identical to the cultured genotype. Found at almost all stations. Highest read abundance along the Hausgarten-Kongsfjorden transect (Figure S5C). Cosmopolitan distribution, rare in the Arctic Basin.
The strains HE492-45, -48, and -52 (Hausgarten) belong to a new clade within the Thalassiosira genus. Cells exclusively solitary, relatively small and elongated, cylindrical with roundish valve edges (Figure 5U). Single, long organic thread extending diagonally from each valve. No matching 28S or 18S rRNA gene sequences in the GenBank. Phylogenetic placement based on 18S rRNA gene suggests that the novel clade is a sister group to T. concaviuscula, though with low bootstrap support (Figure S1). The matching ASV_117 from the metabarcoding dataset found in almost all samples. Most abundant in Hausgarten, outer Kongsfjorden and outer stations of northern fjords (Figure S5D). Arctic-temperate distribution, excluding Arctic Basin.
Family Corethraceae
Corethron sp. strain HE492-01 (Hausgarten). Cells with rounded valves, elliptical appearance in girdle view. Numerous long spines projecting outwards from bases of both valves (Figure 5S). Numerous small round chloroplasts. Only 28S rRNA gene sequence obtained (Figure 7). A single 100% matching sequence in the NCBI, belonging to Corethron sp. (Bay of Fundy). Only one ASV was retrieved from the Corethron clade in the local metabarcoding dataset, which could not be linked with certainty to the cultured strain due to lack of 18S rRNA sequence. The ASV was present in all fjords. Arctic-temperate distribution excluding Arctic Basin (Figure S5E).
Family Leptocylindraceae
Leptocylindrus sp. strain HE492-14 (Hausgarten). Cells cylindrical, elongated, typical morphology for the genus, either solitary or chain-forming (Figure 5R). Numerous small round chloroplasts. Only 28S rRNA sequence obtained. No matching sequences in NCBI (Figure 7). A total of 21 ASVs placed within the Leptocylindrus clade, 20 of which were forming a clade together with L. minimus reference sequences, suggesting either a high undescribed diversity within this genus or a high degree of haplotype diversity within one population. As the 28S sequence of the cultured strain did not place within the L. danicus/L. aporus or L. hargreavesi clades (Figure 7), the strain may belong to the L. minimus clade for which there is no 28S rRNA reference sequence available, but which had a relatively high read abundance in the area. Further studies and 18S rRNA sequencing will be needed to identify this strain and link it with the environmental sequences.
Diatom diversity and distribution in Svalbard fjords based on the metabarcoding data
A total of 230 diatom ASVs were obtained after the processing of the raw metabarcoding data. Following the phylogenetic placement of the ASVs into the newly compiled reference library and the construction of the RAxML tree, 110 ASVs representing 48% of total diatom ASVs could be assigned taxonomically down to the species level with high certainty (Figure S1). The remaining 52% (120 ASVs) could not be placed at the species level due to a lack of reference sequences. On the other hand, genus-level taxonomic identity could be assigned to 87 additional ASVs, thus in total 197 (86%) of diatom ASVs, with only 33 ASVs not being assigned to the genus level. These mainly belonged to different raphid and araphid pennate diatoms (21 ASVs) and polar-centric Mediophyceae (11). Overall, the taxonomic assignment to species level was above average within ecologically important and taxonomically well-studied genera of large centric diatoms such as Chaetoceros, where 16 out of 26 ASVs (62%) were assigned to species level, or Thalassiosira, with 13 out of 19 (68%) ASVs assigned to the species level.
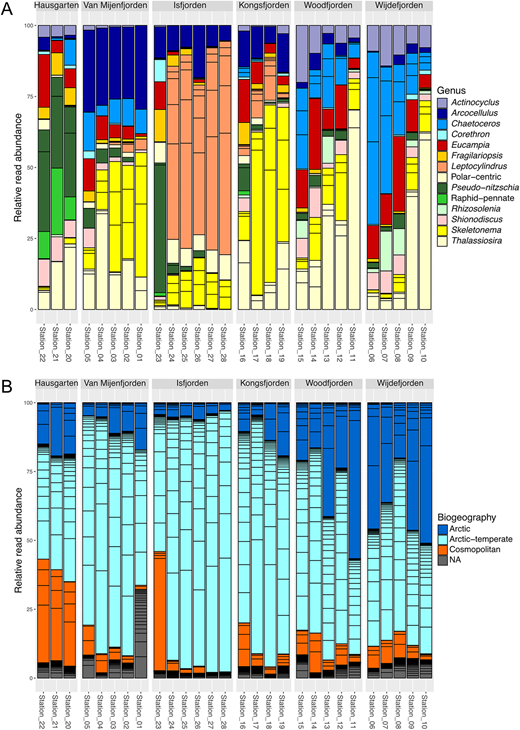
Overall, diatoms had relatively low contribution to the protist community in the studied area based on the metabarcoding data, reaching the maximum of approximately 25% contribution of sequence reads in Isfjorden (Figure S11). Their diversity and distribution patterns, inferred from the metabarcoding data, varied significantly among different fjords (Figures 8A and S12). At Hausgarten stations, the diatom community was dominated by the two Pseudo-nitzschia species (P-n. turgidula and P-n. granii), along with S. bioculatus, Thalassiosira spp. and Eucampia groenlandica. In Van Mijenfjorden, the community was dominated by A. cornucervis, as well as Skeletonema spp. (most notably S. marinoi), Thalassiosira spp. and Chaetoceros spp. The community in Isfjorden was unique within the investigated area, with a high abundance of different ASVs belonging to the L. minimus clade, except at the outermost station, which was dominated by Pseudo-nitzschia spp. A. cornucervis and Skeletonema spp. were also significant components of the Isfjorden community. In Kongsfjorden, the community was primarily dominated by Skeletonema spp., along with A. cornucervis and Thalassiosira spp. Finally, the two northernmost fjords, Woodfjorden and Wijdefjorden, exhibited similar communities dominated by heavily silicified Chaetoceros species (e.g., C. convolutus), Thalassiosira spp. (especially T. hispida), E. groenlandica and Actinocyclus sp.
Diversity and biogeography of diatoms in the Svalbard fjords. (A) Diatom diversity in the studied area shown at the genus level. Only ASVs that constituted more than 10% of total diatom ASVs at each station are included in the plot; (B) Distribution of diatom genotypes in the Svalbard fjords based on their biogeographic type. Each bar segment represents relative abundance of an individual ASV. The ASVs labeled as NA have not been included in the biogeographic analysis.
Diversity and biogeography of diatoms in the Svalbard fjords. (A) Diatom diversity in the studied area shown at the genus level. Only ASVs that constituted more than 10% of total diatom ASVs at each station are included in the plot; (B) Distribution of diatom genotypes in the Svalbard fjords based on their biogeographic type. Each bar segment represents relative abundance of an individual ASV. The ASVs labeled as NA have not been included in the biogeographic analysis.
Biogeographic distribution patterns of the most abundant non-cultured ASVs from the local metabarcoding dataset
To explore the biogeographic patterns of diatoms at the community level using the local metabarcoding dataset, we performed the additional biogeographic analysis of the 25 most abundant ASVs in the dataset that did not correspond to our cultured genotypes (Figures S6–S10; Table S3). When combined with ASVs that matched the cultured strains 100%, these accounted for 97% of total diatom read abundance in the dataset. Of the 25 most abundant non-cultured ASVs, 11 could not be assigned taxonomically at the species level, and one raphid pennate ASV could not be identified at genus level. The biogeographic patterns observed among non-cultured ASVs were similar to those of the cultured strains. The most common biogeographic distribution type was Arctic-temperate (18), followed by Arctic (7). No genotypes from this group exhibited a cosmopolitan distribution.
The ASVs exhibiting Arctic-temperate biogeographic distribution belonged to L. minimus (6 ASVs), E. groenlandica, Thalassiosira concaviuscula, Chaetoceros brevis, Chaetoceros gelidus, Proboscia alata, unidentified species of Actinocyclus, Chaetoceros, Fragilariopsis, Proboscia, Rhizosolenia, Skeletonema and an unknown raphid pennate. The ASVs with Arctic distribution were placed both within well-known Arctic diatom species such as T. hispida, Fragilariopsis cylindrus and C. convolutus, and within unidentified species belonging to genera Skeletonema, Chaetoceros and Bacterosira.
Biogeographic patterns at the community level
The biogeographic types assigned to 20 cultured diatom genotypes (ASVs) and 25 most abundant diatom ASVs from the local metabarcoding dataset were explored at the community level to explore the possible relationship between degree of Atlantic influence and prevalence of biogeographic distribution types in the Svalbard area (Figure 8B). The Hausgarten stations had a unique composition of ASVs, mainly dominated by genotypes with cosmopolitan (approximately 25–40%) and Arctic-temperate (approximately 25–50%) distribution. The prevalence of Arctic genotypes at this station was relatively low (up to 20%). The outermost station in Isfjorden showed a pattern that was similar to Hausgarten stations, with cosmopolitan genotypes accounting for approximately 50% of total read abundance. The relative read abundance of Arctic (up to about 10%) and cosmopolitan (up to 5%) genotypes was generally low in Isfjorden, where Arctic-temperate genotypes made up > 90% of reads. The cosmopolitan genotypes were generally absent from the innermost parts of Isfjorden.
A very similar contribution of different biogeographic types was observed in Van Mijenfjorden and Kongsfjorden. Here, the most abundant group were ASVs with Arctic-temperate distribution (up to approximately 80%), followed by the species that are biogeographically restricted to the Arctic. A slight gradient was observed along these fjords in terms of biogeographic distribution types, with Arctic genotypes increasing, and cosmopolitan genotypes decreasing in contribution towards the innermost stations. Finally, the two northernmost fjords, namely Woodfjorden and Wijdefjorden, had a large proportion of genotypes with Arctic distribution compared with the other studied fjords and Hausgarten stations. The contribution of ASVs restricted to the Arctic was increasing towards the inner parts of the fjords (reaching up to 50% of total read abundance), followed by a decrease in the prevalence of the species with Arctic-temperate and cosmopolitan biogeographic distribution. The species with cosmopolitan distribution had a relatively minor contribution (up to about 10%), and were nearly absent from the innermost stations in Woodfjorden and Wijdefjorden.
Discussion
Bridging the gap between morphological and molecular approaches in studying Arctic diatom diversity
A well-established taxonomic framework, integrating both morphological and molecular approaches, is a prerequisite for studying the composition and structure of Arctic diatom communities and understanding how they shape the Arctic ecosystem function. In this work, we used morphological and molecular characterisation of diatom cultures to link robust taxonomic identification to local and global metabarcoding datasets. The precise identification at the species and genotype level allowed for detecting general biogeographic patterns that are underlying diatom community structure in the Svalbard area.
Large-scale cultivation efforts are crucial for bridging the gap between traditional morphology-based identification of diatoms and state-of-the-art molecular metabarcoding approaches. The phylogenetic analysis of cultured and environmental genotypes shows that Arctic diatom diversity is not yet fully represented in morphological and molecular databases, as demonstrated in recent large-scale diatom cultivation studies in the Arctic (Balzano et al., 2017; Ribeiro et al., 2020). Here, we have found seven diatom strains that could not be identified to species level using light microscopy or available molecular reference libraries. Probable novel species were found even within the taxonomically well studied planktonic genera such as Chaetoceros and Thalassiosira. We also found a cryptic species and a novel sub-clade within a globally distributed C. diadema species complex that is biogeographically restricted to the Arctic realm. These findings underline the importance of further taxonomic studies based on molecular methods and suggest that Arctic diatom communities harbour a significant undescribed diversity of species or genotypes that are specifically adapted to the extreme Arctic environment and endemic to the Arctic.
The taxonomy assignment of ASVs from the local metabarcoding datasets confirmed the notion that the interpretative power of metabarcoding is directly linked to the representation of taxa in reference libraries (Zimmermann et al., 2015; Gaonkar et al., 2020). In our analysis, about half of the diatom ASVs could be identified to the species level, and this level of identification was only possible for groups with high-quality reference sequences such as the genus Chaetoceros (Gaonkar et al., 2018) or other well-studied planktonic genera like Pseudo-nitzschia and Thalassiosira. Overall, Arctic diatoms are underrepresented in reference databases, and large-scale cultivation is needed to link morphological and molecular information. Notable gaps in reference databases could be observed for pennate diatoms, where a significant portion of the ASVs could not be identified to species or even genus level. In particular, our study did not include sea-ice or benthic communities, where pennate diatoms are especially ecologically relevant and diverse, and where gaps in reference databases are likely to be even higher. The 23 cultured genotypes accounted for about a third (35%) of the total diatom sequence reads in the metabarcoding dataset, showing that combining cultivation with metabarcoding surveys can significantly improve the subsequent interpretation of metabarcoding data.
We used two common genetic markers for species-level identification and phylogenetic placement of diatom cultures, namely the 18S and 28S rRNA gene sequences. The two showed overall similar taxonomic resolution and proved to be sufficient for discerning different diatom morphotypes, as was demonstrated earlier (Zimmermann et al., 2015; Gaonkar et al., 2020). The clear advantage of 18S rRNA, apart from being a well-established marker for metabarcoding of diatoms (Zimmermann et al., 2011), is in the availability of reference databases such as PR2 (Guillou et al., 2013) and of large metabarcoding datasets such as Tara Ocean (de Vargas et al., 2015), the Ocean Sampling Day (Kopf et al., 2015) and Malaspina (Duarte, 2015) that allow for global biogeographic mapping of ASVs, as shown here and in similar studies on diatoms (De Luca et al., 2019) and other protists (Ichinomiya et al., 2016; Lopes dos Santos et al., 2017). In some cases, the 18S rRNA marker was not sufficient for discerning morphospecies, as was shown for the two strains of C. debilis that could only be separated using the 28S rRNA sequences. Similarly, the T. gravida cultures had identical 18S and partial 28S rRNA gene sequences to T. rotula sequences, as the two can only be discerned using more variable genes such as ITS rRNA (Whittaker et al., 2012).
In this study, the V4 region of the 18S rRNA gene marker appeared to have sufficient resolution for species-level identification of diatoms in most cases. This sufficiency means that the local and global compiled 18S rRNA metabarcoding datasets can be used to determine biogeographic distribution patterns at the genotype level, likely reflecting the ecological niche of a genotype. Clearly, the use of more variable markers such as ITS rRNA, rbcL and psbC that can differentiate additional diatom genotypes may reveal finer biogeographic patterns. However, metabarcoding datasets based on these markers are lacking, and 18S rRNA gene datasets presently provide the best global coverage.
In some cases, we found a high number of ASVs within certain terminal diatom clades. This finding was most notable within the L. minimus clade, but also within other clades such as C. contortus, A. cornucervis or the new strain from the Cymatosiraceae family. This high diversity of ASVs may indicate high intragenomic haplotype diversity of the 18S rRNA gene, as was shown in environmental metabarcoding data from the genus Chaetoceros (Gaonkar et al., 2020) and within clonal strains of the genus Chaetoceros (De Luca et al., 2021), where dominant haplotype is accompanied with peripheral haplotypes with order of magnitude lower read numbers. High intragenomic haplotype diversity was also reported for the genus Skeletonema (Alverson and Kolnick, 2005). Another argument for the common genomic origin of dominant and peripheral ASVs within L. minimus and other clades is the similarity of distribution patterns among the most abundant ASVs in L. minimus clade, both within the local dataset and globally. In light of these observations, applying a 100% pairwise identity when linking cultures with metabarcoding datasets likely reveals only the patterns of the dominant haplotype which is also the one obtained by Sanger sequencing of cultures. The dominant haplotype is nevertheless highly representative of a studied genotype, and the analysis is not likely losing significant biogeographic information from minor haplotypes. On the other hand, differences in the global distribution among some of the peripheral ASVs would suggest that they indeed represent distinct species that are currently lacking in the reference databases.
Biogeographic patterns of Arctic diatoms
Marine planktonic protists, including diatoms, have generally been considered cosmopolitan (globally distributed), with unlimited potential for dispersal through oceanic circulation, and their community structure primarily regulated by environmental forcing (Finlay, 2002). In particular, the population structure of marine planktonic diatoms has been argued to be defined almost exclusively by environmental gradients with no long-term limitations to dispersal that could result in biogeographically restricted or endemic planktonic diatom taxa (Cermeño and Falkowski, 2009). Despite recent findings suggesting that endemic diatoms are common in freshwater systems (Vanormelingen et al., 2009), notably in polar regions (Vyverman et al., 2010), there has been no strong evidence for endemism in marine planktonic diatoms. Still, early morphology-based studies (Hasle, 1976) have shown that planktonic diatoms can exhibit certain biogeographic patterns such as cold-water, warm-water, and cosmopolitan distribution.
In recent years, findings of cryptic cold-adapted, planktonic diatom species within presumably cosmopolitan species complexes (Chamnansinp et al., 2013; Percopo et al., 2016) have suggested that the morphology-based taxonomy and morphology-based datasets may, in many cases, prove insufficient for detecting biogeographic patterns and potential endemism or restricted distribution in diatoms. Integrating morphology-based records with global molecular metabarcoding datasets, such as Tara Ocean (Karlusich et al., 2020) and Ocean Sampling Day (Kopf et al., 2015), revealed that genotypes of Chaetoceros show both endemic and biogeographically restricted distribution patterns, notably in polar areas (De Luca et al., 2019). Building on this research, we demonstrate here that genotypes of Arctic planktonic diatoms exhibit a variety of biogeographic patterns, ranging from endemic (or Arctic-restricted distribution) to cosmopolitan distribution.
After analysing biogeographic patterns of 45 diatom genotypes from the Svalbard area, we detected three distinct general biogeographic distribution types, namely Arctic, Arctic-temperate and cosmopolitan. Overall, there was no clear link between phylogeny and biogeography, and species within the most relevant diatom genera such as Chaetoceros and Thalassiosira exhibited a wide range of biogeographic distribution types. These distribution types generally correspond well with the global biogeographic distribution of Arctic diatoms defined by Balzano et al. (2017) and show some similarities with the earlier study of Hasle (1976), both of which used morphology-based biogeographic analyses. For example, the biogeographic patterns delineated by Balzano et al. (2017) and this study were the same for A. cornucervis (Arctic-temperate), C. closterium (cosmopolitan), E. groenlandica (Arctic-temperate), T. gravida (Arctic-temperate), C. gelidus (Arctic-temperate) and C. neogracilis (Arctic-temperate). On the other hand, slightly different biogeographic distribution was observed in this study compared with that of Balzano et al. (2017) for P.-n. granii (cosmopolitan in this study versus Arctic-temperate), T. hispida (Arctic versus Arctic-temperate), S. bioculatus (cosmopolitan versus Arctic-temperate). Observed differences in biogeography of individual species between this study and solely morphology-based studies can have different causes. They may stem from the finer, genotype-level approach used here that could distinguish cold-adapted cryptic species, but also in some cases from insufficient resolution of 18S rRNA marker when distinguishing morphologically distinct but closely related species, as is often the case in the Pseudo-nitzschia genus (Lundholm et al., 2012).
The most common biogeographic distribution type among the studied genotypes was Arctic-temperate, which included 30 genotypes that are present both at temperate latitudes (30–66oN) and above the polar circle (66oN). Within this general biogeographic group that was dominating diatom communities in the fjords on the west coast of Svalbard (at Hausgarten, Van Mijenfjorden, Isfjorden and Kongsfjorden), we observed a range of possible finer biogeographic patterns that were difficult, however, to delineate objectively based on 18S V4 rRNA metabarcoding datasets used in this study. For example, some Arctic-temperate genotypes (e.g., C. neogracilis and A. longicornis) have clearly established communities in the Arctic Basin (beyond the polar front) and are also found in sea-ice communities. On the other hand, Arctic-temperate genotypes such as C. contortus, Corethron sp., or some genotypes of L. minimus, are common in the Svalbard area, but are not present beyond the polar front. These Arctic-temperate genotypes, absent from the Arctic Basin, are of particular interest as they are seemingly mainly associated with relatively warmer Atlantic waters (> 3 oC), and their northwards expansion can indicate Atlantification or ocean warming in the area.
Similar patterns could be observed among the cosmopolitan genotypes. Out of the five cosmopolitan genotypes, only two were present continuously at all latitudes between the Arctic Basin and Antarctica (S. bioculatus, and C. closterium). The remaining three, P.-n. granii, P.-n. turgidula and T. oceanica, were generally absent from the Arctic Basin. In particular, P.-n. turgidula was mostly found at Hausgarten, suggesting that it represents a warm-water genotype associated with Atlantic waters. These observations also indicate that finer biogeographic patterns and additional cryptic diversity would likely be revealed for these genotypes using finer genetic markers, as demonstrated by detection of cold-adapted C. closterium ecotypes based on 28S rRNA gene (Stock et al., 2019).
Ten genotypes were endemic to the Arctic. These belonged to species which have already been considered as restricted to the Arctic (and sometimes to southern polar areas as well) based on morphology-based studies, such as C. convolutus and F. cylindrus (Lundholm and Hasle, 2010; Balzano et al., 2017), but also to some cryptic genotypes within species complexes with wider biogeographic distribution such as C. diadema, T. hispida and S. costatum. This work demonstrates that biogeographically restricted distribution or Arctic endemism not only exists but is relatively common among planktonic diatoms in the Svalbard area. Similar studies of sea ice-associated diatom genotypes would undoubtedly reveal additional endemism among Arctic diatoms, which has currently been suspected or assumed for a number of sea ice-associated pennate diatoms (Quillfeldt, 2000; Poulin et al., 2011).
Examination of biogeographic distribution types on the community level within five Svalbard fjords and Hausgarten area revealed distinct patterns in the abundance of genotypes with specific distribution types. Overall, diatom communities were shaped by the three main groups of genotypes: 1) genotypes restricted to the Arctic, that dominated in the northernmost fjords (Woodfjorden and Wijdefjorden); 2) genotypes with Arctic-temperate distribution, that dominated fjords on the west coast of Svalbard (Van Mijenfjorden, Isfjorden and Kongsfjorden), and 3) genotypes with cosmopolitan distribution, that dominated at Hausgarten stations. Within groups 2 and 3, there seemed to exist additional distribution patterns, with differing degrees of expansion within the Arctic realm. This finding suggests that the diatom communities in the area are structured by an interplay of several processes: dispersal and advection of warm water genotypes and their ability to survive the extreme Arctic environment (especially beyond the polar front), ability of genotypes restricted to the Arctic to expand southwards into warmer waters, and an underlying stock of genotypes which seem to be well-adapted both to warmer waters and the Arctic.
Advection is a well-documented source of highly productive, warm, and nutrient-rich Atlantic waters into Fram Strait and the western Svalbard area (Randelhoff et al., 2015; Wassmann et al., 2015; Wassmann et al., 2019). An important component of advection is the transport of planktonic organisms into the Arctic realm, a phenomenon that has been well-documented for temperate mesozooplankton (Wassmann et al., 2019). To date, the main evidence for the transport of phytoplankton with Atlantic waters was the northward expansion of the haptophyte, Emiliania huxleyi, reported in several studies from Fram Strait and the Barents Sea (Hegseth and Sundfjord, 2008; Neukermans et al., 2018; Oziel et al., 2020). Another example is the northward expansion in the Atlantic Arctic of the cyanobacterium Synechoccocus (Paulsen et al., 2016). The results from this study demonstrate for the first time that diatom genotypes with broad Arctic-temperate and cosmopolitan distribution are abundant in the Svalbard area, especially in the areas with stronger Atlantic influence.
While the local metabarcoding data shown here provide only a snapshot of the late Arctic summer diatom community, they clearly demonstrate that species with wider biogeographic distribution dominated the Atlantic-influenced waters, and genotypes endemic to the Arctic dominated colder waters. Despite the clear general trend, the finer scale genotype-level patterns provided a much more complex picture. For example, the cosmopolitan P.-n. turgidula genotype was not detected in Van Mijenfjorden and was nearly absent from the northernmost fjords, while another cosmopolitan genotype P.-n. granii was common in all fjords. Moreover, while the majority of the Arctic-temperate genotypes were present in all fjords, most of the Arctic-temperate L. minimus genotypes were nearly absent from the innermost parts of the northern fjords. On the same note, the F. cylindrus genotype, endemic for the Arctic, was uniformly distributed throughout the area. These patterns point out that genotype-level ecophysiology is also important for shaping diatom community structure, as it likely determines the local distribution and abundance of individual genotypes with respect to specific hydrography of each fjord and seasonal variation in environmental drivers (Basedow et al., 2018; Menze et al., 2020).
Conclusion
This work underscores the importance of combining morphological and molecular approaches in studying biodiversity and biogeography of Arctic diatoms. By providing novel morphology-based data and taxonomic descriptions as well as novel reference sequences of 18S and 28S rRNA marker genes, it represents an important contribution to future studies on Arctic diatom biodiversity. Moreover, we demonstrated that Arctic diatom flora harbours a significant undescribed biodiversity, and that new taxa are also found within morphologically well-described genera of planktonic diatoms. Future studies should focus on formally describing these new taxa and expanding on the intensive cultivation efforts to further reveal the biodiversity of Arctic diatoms. Our biogeographic analysis shows that diatom genotypes from the Svalbard area exhibit a range of biogeographic patterns, from endemic genotypes restricted to the Arctic, to genotypes with wider, Arctic-temperate and cosmopolitan biogeographic distribution. Using finer taxonomic markers (e.g., ITS rDNA), capable of delineating cryptic Arctic species and cold-adapted Arctic genotypes, would likely reveal additional, finer scale biogeographic patterns, providing better insight into biogeographic processes that are shaping the Arctic community structure.
Data accessibility statement
CTD data from the cruise is available on Pangea link: John and Wisotzki (2017): Physical oceanography during HEINCKE cruise HE492; https://doi.org/10.1594/PANGAEA.881306. Raw Ilumina metabarcoding data from the HE492 cruise have been deposited to GenBank SRA under accession number: PRJEB49358. Sequences of the strains are available at NCBI with accession numbers OK147649-147680 for 18S rRNA sequences, and OK147681-147729 for 28S rRNA sequences (Table 1). Scripts and processed data are available from https://github.com/vaulot/metapr2_HE492.
Supplemental files
The supplemental files for this article can be found as follows:
Figure S1 is provided both as a supplemental file and as an interactive phylogenetic tree in ITOL: https://itol.embl.de/tree/193157255105100381657613640. Supplementary PDF document including 11 supplementary figures (Figure S2–S12) and three supplementary tables (Table S1–S3) is submitted with the document.
Acknowledgments
We are thankful to the crew on board R/V Heincke for their assistance during the HE492 cruise in Svalbard fjords. Jan Heuschele (UiO) provided valuable help with coding in R and data analysis. Bioinformatic analyses were performed on the UiO UNINETT Saga2 computer cluster, project nn9307k, with help from Anders Kristian Krabberød (UiO). We thank the Roscoff ABIMS platform of the FR2424 (CNRS, Sorbonne Université) for bioinformatics resources.
Funding
This study was financially supported by The Research Council of Norway (RCN) to the projects “TaxMArc: Taxonomy of marine protists in a changing Arctic” no. 268286 (LS, BE, UJ, DV) and by ERA-MBT initiated project “PROMiSE” (SGS and BE no. 282936). UJ was supported by the “PROMiSE” project no. 161B0561A from DLR/BMBF. UJ and KK were supported by ERA4CS, an ERA-NET initiated by JPI Climate, and funded DLR/BMBF (DE): 01LS1714A with co-funding by the European Union (Grant 690462). UJ was DFG supported to SPP1991, “Taxonomics” project GZ: JO 702/13-1 AOBJ: 641030. UJ received funding and support for the HE492 cruise within the Helmholtz research program “Changing Earth, Sustaining our Future” (Sub-topic 6.2 Adaptation of marine life) of the Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research, Germany. DV was supported by ANR contract “PhytoPol” (ANR-15-CE02-0007).
Competing interests
The authors have declared that no competing interests exist.
Author contributions
Contributed to conception and design: LS, BE, UJ.
Contributed to acquisition of data: LS, KK, SGS, CH, BE, UJ.
Contributed to analysis and interpretation of data: LS, SGS, DV, BE, UJ.
Drafted and/or revised the article: LS, KK, SGS, CH, DV, BE, UJ.
Approved the submitted version for publication: LS, KK, SGS, CH, DV, BE, UJ.
References
How to cite this article: Šupraha, L, Klemm, K, Gran-Stadniczeñko, S, Hörstmann, C, Vaulot, D, Edvardsen, B, John, U. 2022. Diversity and biogeography of planktonic diatoms in Svalbard fjords: The role of dispersal and Arctic endemism in phytoplankton community structuring. Elementa: Science of the Anthropocene 10(1). DOI: https://doi.org/10.1525/elementa.2021.00117
Domain Editor-in-Chief: Jody W. Deming, University of Washington, Seattle, WA, USA
Associate Editor: Kevin R. Arrigo, Department of Earth System Science, Stanford University, Stanford, CA, USA
Knowledge Domain: Ocean Science